
A connectome in the mouse visual cortex covering all the layers
Resources
-

EM Images
-

Dense Segmentation
-

Excitatory Neurons
-

Inhibitory Neurons
-
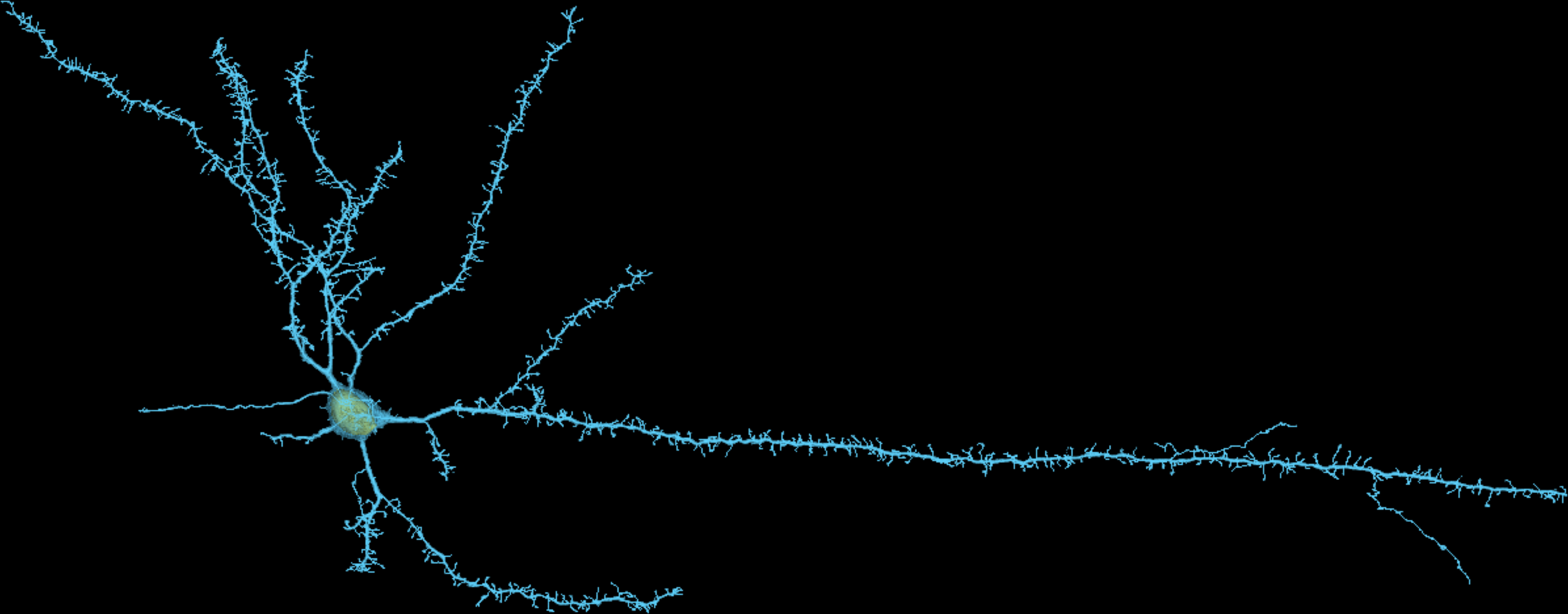
Neuron with Neucleus
-

Synapse
Access
EM Images: s3://bossdb-open-data/iarpa_microns/basil/em
Segmentation/Meshes/Skeletons: s3://bossdb-open-data/iarpa_microns/basil/seg/
Synaptic Clefts: s3://bossdb-open-data/iarpa_microns/basil/clefts/
Nuclei: s3://bossdb-open-data/iarpa_microns/basil/nuclei/
Connectome Graph: https://bossdb-open-data.s3.amazonaws.com/iarpa_microns/basil/synapse_graph/cleaned_final.csv.gz
Contributions
Co-registration: Nuno Maçarico da Costa, Adam Bleckert, Marc Takeno
Tissue Preparation: Joann Buchanan, Marc Takeno, Nuno Maçarico da Costa
Sectioning: Agnes Bodor, Adam Bleckert
TEM Design and Maintenance: Derrick Brittain, Clay Reid
TEM Operation: Derrick Brittain, Wenjing Yin, Adam Bleckert, Marc Takeno, Daniel Bumbarger, Nuno Da Costa
EM Stitching and Rough Alignment: Gayathri Mahalingam, Russel Torres, Yang Li, Thomas Macrina, Dodam Ih
EM Fine Alignment: Thomas Macrina, Will Wong
Neuron segmentation: Kisuk Lee, Jingpeng Wu
Agglomeration: Ran Lu
Synapse Detection: Nicholas Turner, Jingpeng Wu
Neuroglancer Frontend: Nico Kemnitz, Manuel Castro
Proofreading Backend: Sven Dorkenwald, Nico Kemnitz, Chris S. Jordan
Cloud Data Interface: William Silversmith, Ignacio Tartavull
Connectome Versioning System: Forrest Collman, Sven Dorkenwald, Casey Schneider Mizell
Proofreading: Agnes Bodor, Nuno Maçarico da Costa, Szi-Chieh Yu, Alyssa Wilson, Sven Dorkenwald, Casey Schneider-Mizell, Forrest Collman
Project Management: Lynne Becker, Shelby Suckow
Scientific Management: Clay Reid, Nuno Da Costa, Andreas S. Tolias, Jacob Reimer, H. Sebastian Seung
IARPA MICrONS Program Management: David A. Markowitz, Jacob Vogelstein
Publications
Under preparation